SPANDx workflow for analysis of haploid next-generation re


PDF) SPANDx: A genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets

Agronomy, Free Full-Text
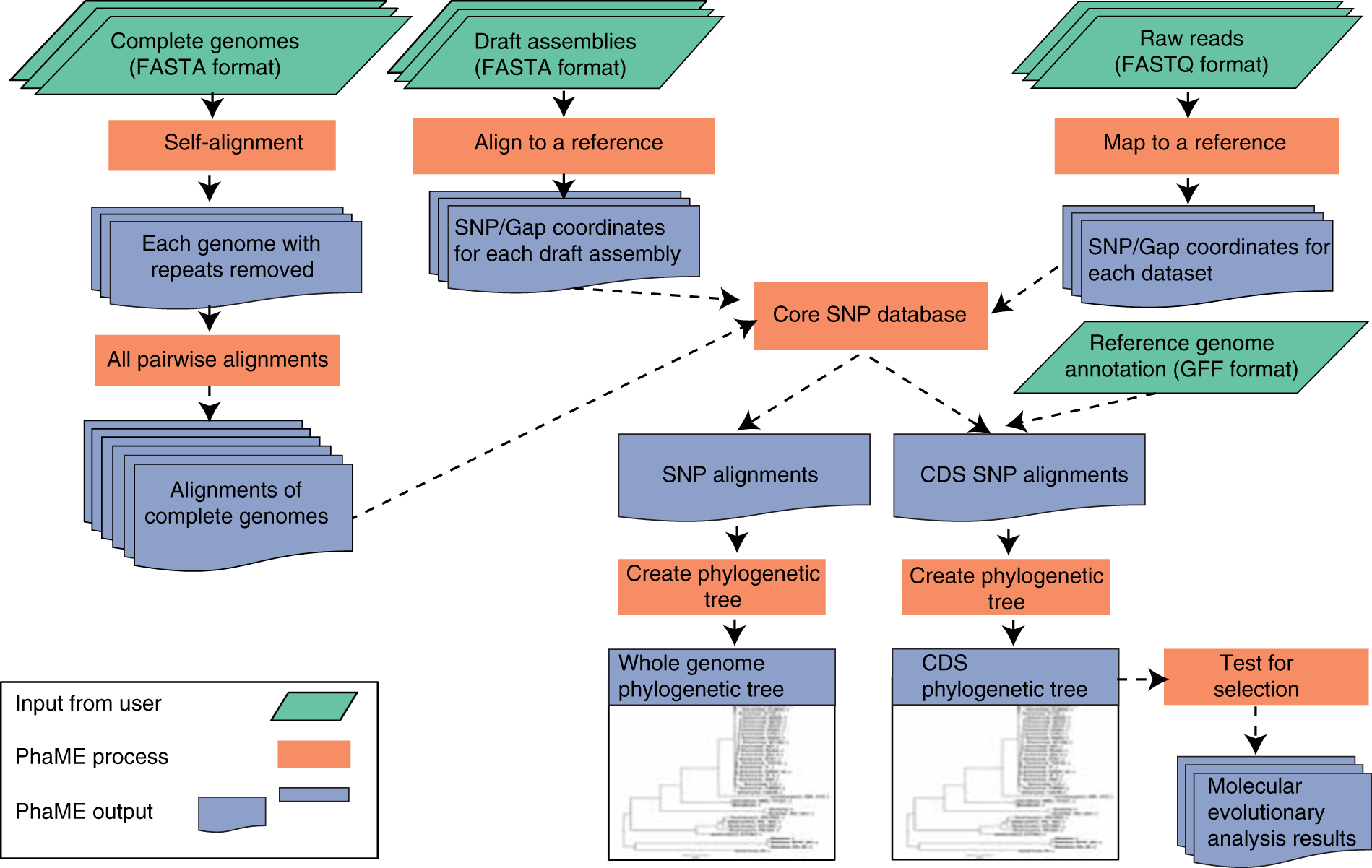
Standardized phylogenetic and molecular evolutionary analysis applied to species across the microbial tree of life

Save time with our One-Day-Workflow - GenDx

Derek SAROVICH, Senior Research Fellow, PhD Microbiology, University of the Sunshine Coast, USC, Faculty of Science, Health, Education and Engineering

SPANDx workflow for analysis of haploid next-generation re-sequencing data.
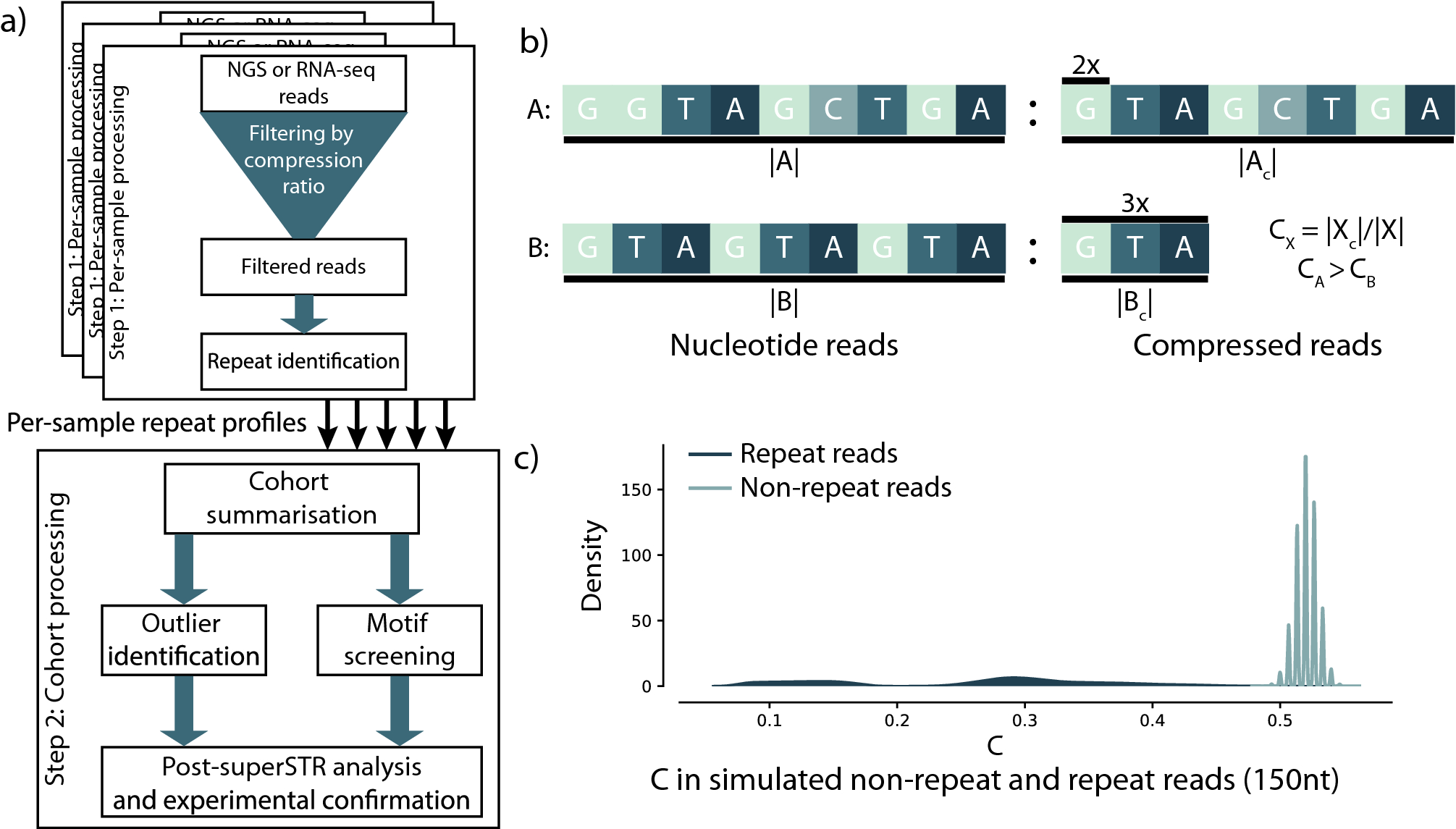
Detection of repeat expansions in large next generation DNA and RNA sequencing data without alignment

SPANDx workflow for analysis of haploid next-generation re-sequencing data.
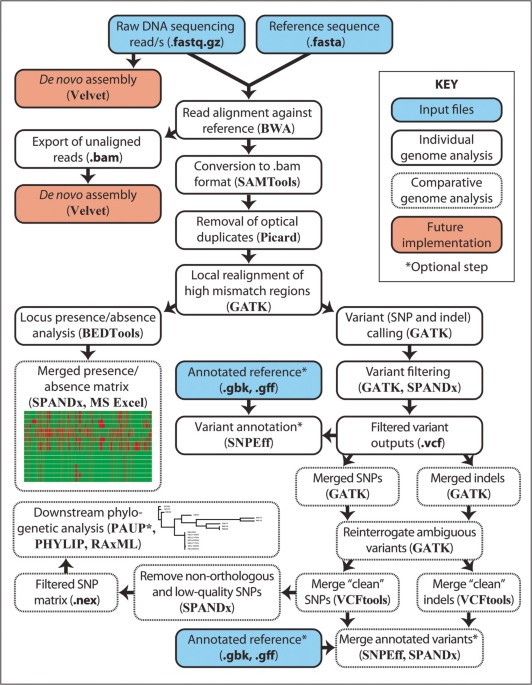
SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets, BMC Research Notes

SPANDx workflow for analysis of haploid next-generation re-sequencing data.

Derek SAROVICH, Senior Research Fellow, PhD Microbiology, University of the Sunshine Coast, USC, Faculty of Science, Health, Education and Engineering

SPANDx workflow for analysis of haploid next-generation re-sequencing data.

SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets, BMC Research Notes